Our Local Distributor
北京 101111
Quick Overview for CUT&RUN Pro Set (ABIN6923138)
应用范围
-
-
适用
- Eukaryotes
-
原理
- This set contains Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays, CUT&RUN Positive and Negative Control for the CUT&RUN method for improved genome-wide detection of Protein-DNA-Interactions.
-
产品特性
-
CUT&RUN (Cleavage Under Targets And Release Using Nuclease) offers a new approach to pursue epigenetics.
CUT&RUN overcomes various downfalls of ChIP-Seq with improved workflow.
CUT&RUN-Sequencing has the advantage of being a simpler technique with lower costs due to the high signal-to-noise ratio, requiring less depth in sequencing.
CUT&RUN has the potential to replace all ChIP-based applications. -
组件
-
- CUT&RUN Positive Control (Recombinant Rabbit anti-H3K27me3 Antibody)
- CUT&RUN Negative Control (Polyclonal Guinea Pig anti-Rabbit IgG Antibody, Pre-Adsorbed)
- Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays
-
试剂未包括
-
- Specific antibody against target of interest
- pA/G-MNase (ABIN6950951)
-
-
-
-
试剂准备
-
- Wash Buffer
- Binding Buffer
- Antibody Buffer
- Digitonin Wash Buffer
- 2x Stop Buffer
- Low Salt Rinse Buffer
- Low Salt Incubation Buffer
- Low Salt Stop Buffer
-
实验流程
-
Cell Harvest
- Harvest cells for each sample at RT
- Wash cells 4 x with 1 mL Wash Buffer
Prepare Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays
- Pipette 10 µL Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays slurry for each sample into a tube
- Place the tubes on a magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Wash beads 3 more times with 1 mL Binding Buffer
- Finally resuspend the beads with 10 µL Binding Buffer per sample
Cell Immobilization – binding to Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays
- Carefully vortex the samples and add 10 µL of the prepared Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays to each sample
- Close tubes tightly and rotate for 5-10 min at RT
Cell Permeabilization and Primary Antibody Binding
- Place the tubes on a magnetic separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Place each tube on the vortex mixer set to a low speed and add 100 µL Antibody Buffer containing Digitonin
- Gently vortex the tubes until the beads are resuspended
- Add 5 µL CUT&RUN anti-DYKDDDDK antibody or CUT&RUN Positive Control or CUT&RUN Negative Control corresponding to a 1:20 dilution
- Add 1 µL primary antibody against your protein of interest corresponding to a 1:100 dilution to the remaining samples
- Rotate the tubes for 2 h at RT or 4 h to O/N at 4 °C
- Place the tubes on a magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Resuspend pellet with 1 mL Digitonin Wash Buffer and mix by inversion
- Wash again
Secondary Antibody Binding (optional)
If no secondary antibody is used proceed directly to pA/G-MNase-Binding.- Place the tubes on a magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Vortex the samples at low speed and add 100 µL Digitonin Wash Buffer per sample
- Add 5 µL Secondary Antibody corresponding to a 1:20 dilution
- Rotate the tubes for 1 h at 4 °C
- Place the tubes on a magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Resuspend with 1 mL Digitonin Wash Buffer and mix by inversion
- Wash again
Protein A-MNase or Protein A/G-MNase Binding
- Place the tubes on a magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Place each tube on the vortex mixer set to a low speed and add 50 µL Digitonin Wash Buffer and 2.5 µL pA/G-MNase per sample
- Rotate the tubes for 1 h at 4 °C
- Place the tubes on a magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Resuspend with 1 mL Digitonin Wash Buffer and mix by inversion
- Wash again
MNase Digestion and Release of pA/G-MNase-Bound Chromatin Fragments
High Ca2+/Low Salt Chromatin Cleavage
- Quick pulse in a table-top centrifuge (max 100 x g)
- Place the tubes on a magnet separator and remove the liquid carefully
- Resuspend with 1 mL Low-Salt Rinse Buffer and mix by inversion
- Quick pulse in a table-top centrifuge (max 100 x g)
- Place the tubes on a magnet separator and remove the liquid carefully
- Wash again
- Place each tube on the vortex mixer set to a low speed and add 200 µL ice cold Low Salt Incubation Buffer per sample
- Incubate tubes at 0 °C for 5 min
- Place the tubes on a cold magnet separator and remove the liquid carefully
- Remove the tubes from the magnetic separator
- Resuspend with 200 µL Low Salt Stop Buffer and mix by gentle vortexing
- Incubate tubes at 37 °C for 30 min
- Place the tubes on a magnet separator
- Transfer the supernatant containing the pA/G-MNase-bound digested chromatin fragments to fresh 1.5 mL tubes
- Proceed with DNA extraction
-
限制
- 仅限研究用
-
-
- by
- New strategies to inhibit tumor angiogenesis laboratory headed by Prof. Elisabetta Dejana, IFOM - the FIRC institute of Molecular Oncology
- No.
- #104253
- 日期
- 2021.02.18
- 抗原
- H3K27me3
- Lot Number
- CR0109190001
- Method validated
- Cleavage Under Targets and Release Using Nuclease
- Positive Control
- Recombinant rabbit anti-H3K27me3 antibody
- Negative Control
- Polyclonal guinea pig anti-rabbit IgG antibody
- Notes
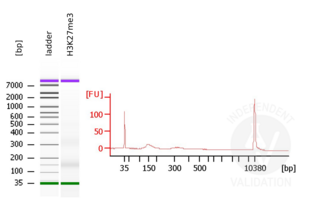
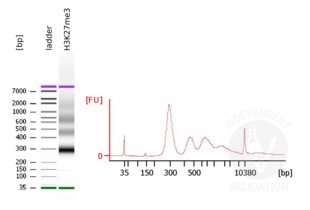
Passed. ABIN6923144 is suitable for CUT&RUN to prepare H3K27me3 targeted DNA fragments from genomic murine DNA.
- Primary Antibody
- Secondary Antibody
- Full Protocol
- Cell harvest
- Harvest 300,000 murine endothelial cells for each sample at RT. Keep cells for each sample in separate tubes.
- Centrifuge cell solution 3 min at 600 x g at RT.
- Remove the liquid carefully.
- Gently resuspend cells in 1 ml Wash Buffer by pipetting and transfer cell solution to a 1.5 ml microcentrifuge tube.
- Centrifuge cell solution 3 min at 600 x g at RT and discard the supernatant.
- Repeat three times for a total of four washes.
- Resuspend cell pellet for each sample in 1 ml Wash Buffer by gently pipetting.
- Concanavalin A beads preparation
- Prepare one 1.5 ml microcentrifuge tube for each sample.
- Gently resuspend the CUT&RUN Concanavalin A Beads.
- Pipette 10 µl CUT&RUN Concanavalin A Beads slurry for each sample into the 1.5 ml microcentrifuge tubes.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Pipette 1 ml Binding Buffer into each tube and resuspend CUT&RUN Concanavalin A Beads by gentle pipetting.
- Spin down the liquid from the lid with a quick pulse in a bench-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Repeat twice for a total of three washes.
- Gently resuspend the CUT&RUN Concanavalin A Beads in a volume of Binding Buffer corresponding to the original volume of bead slurry, i.e. 10 µl per sample.
- Cell immobilization – binding to Concanavalin A beads
- Carefully vortex the cell suspension and add 10 µl of the CUT&RUN Concanavalin A Beads in Binding Buffer to each sample.
- Close tubes tightly and rotate for 10 min at RT.
- Cell permeabilization and primary antibody binding
- Place the microcentrifuge tubes on a magnetic stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Place each tube at a low angle on the vortex mixer set to a low speed and add 100 µl Antibody Buffer containing digitonin.
- Gently vortex the microcentrifuge tubes until the beads are resuspended.
- Add 2 µl H3K27me3 positive control antibody ABIN6923144 corresponding to a 1:50 dilution.
- Rotate the microcentrifuge tubes for O/N at 4 °C.
- Spin down the liquid and place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 1 ml Digitonin Wash Buffer and mix by inversion. If clumps occur, gently remove the clumps with a 1 ml pipette tip.
- Repeat once for a total of two washes.
- pAG-MNase Binding
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Vortex the sample at low speed and add 50 μl Digitonin Wash Buffer per sample along the side of the tube. Add 2.5µl CUTANA™ pAG-MNase for ChIC/CUT&RUN Assays (ABIN6950951, lot 19199003).
- Rotate the microcentrifuge tubes for 1 h at 4 °C.
- Spin down the liquid and place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 1 ml Digitonin Wash Buffer and mix by inversion. If clumps occur, gently remove the clumps with a 1 ml pipette tip.
- Repeat once for a total of two washes.
- MNase digestion and release of pAG-MNase-antibody-chromatin complexes
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Resuspend with 1 ml Low Salt Rinse Buffer and mix by inversion. If clumping occurs, gently remove the clumps with a 1 ml pipette tip.
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Repeat once for a total of two washes.
- Place each tube at a low angle on the vortex mixer set to a low speed and add 200 μl ice cold Low Salt Incubation Buffer per sample along the side of the tube.
- Incubate tubes at 0 °C for 1 h.
- Place the tubes on a cold magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Resuspend with 200 µl Low Salt Stop Solution and mix by gentle vortexing.
- Incubate tubes at 37 °C for 30 min.
- Place the tubes on a magnet stand until the fluid is clear.
- Transfer the supernatant containing the pAG-MNase-bound digested chromatin fragments to fresh 1.5 ml microcentrifuge tubes.
- DNA extraction
- Add 2 µl 10% SDS to a final concentration of 0.1% and 5 µl Proteinase K (10 mg/ml) to a final concentration of 0.25 mg/ml to each supernatant containing the pAG-MNase-bound digested chromatin fragments.
- Gently vortex tubes at a low speed of approximately 1100 rpm.
- Incubate tubes at 37 °C O/N.
- Add 200 µl Phenol-Chloroform-Isoamyl alcohol (PCI) to tube.
- Vortex tubes thoroughly at high speed until the liquid appears milky.
- Transfer liquid to a phase-lock tube.
- Centrifuge tubes in a tabletop centrifuge at 16000 x g at RT for 5 min.
- Carefully transfer the upper aqueous phase to a fresh 1.5 ml microcentrifuge tube containing 200 µl chloroform:isoamyl alcohol 24:1 solution.
- Vortex tubes thoroughly at high speed until the liquid appears milky.
- Centrifuge tubes in a benchtop centrifuge at 16000 x g at 4 °C for 5 min.
- Carefully transfer to upper aqueous phase to a fresh 1.5 ml microcentrifuge tube containing 2 µl glycogen (diluted 1:10 to 2 mg/ml from the 20 mg/ml stock solution).
- Add 20 µl 3 M NaOAc pH 5.2 or 150 µl 5 M NH4OAc.
- Add 500 µl 100% ethanol.
- Place O/N at -20 °C.
- Centrifuge tubes in a benchtop centrifuge at 16000 x g at 4 °C for 5min.
- Remove the liquid carefully with a pipette.
- Add 1ml 70% ethanol.
- Centrifuge tubes in a benchtop centrifuge at 16000 x g at 4 °C for 1 min.
- Remove the liquid carefully with a pipette.
- Air-dry the pellet, then dissolve in 15 µl 1 mM Tris-HCl, 0.1 mM EDTA.
- Sequencing library preparation
- Sequencing libraries were prepared using the KAPA HyperPrep Kit (Kapa Biosystems, KR0961) according to the manufacturer’s recommendations with the following modification:
- For the post-ligation cleanup kit, the SPRI bead to ligation reaction ratio was increased to 1.1 to avoid loss of CUT&RUN products.
- The PCR conditions were optimized for short products to avoid melting of the small fragments during elongation and favor short PCR products:
- Initial denaturation: 1 cycle: 45 sec at 98 °C
- Amplification: 16 cycles: 15 sec at 98 °C, followed by 10 sec at 60 °C
- Final extension: 1 cycle for 1 min at 72°C
- Sample quality control
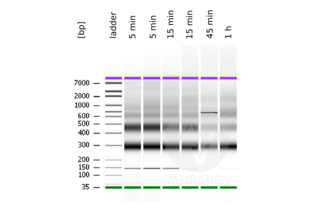
- Evaluate DNA fragmentation via Bionanalyzer Electrophoresis before and after library preparation.
- Experimental Notes
MNase digestion was tested for 5 min, 15 min, 45 min, and 1 h. DNA from the 1 h digestion reaction was selected for library preparation because of the higher ratio of mononucleosomal fragments.
生效 #104253 (Cleavage Under Targets and Release Using Nuclease)![成功验证 '独立验证'标志]()
![成功验证 '独立验证'标志]() Validation ImagesFull Methods
Validation ImagesFull Methods -
-
缓冲液
-
CUT&RUN Positive Control: 50 % Glycerol/PBS, 1 % BSA, 0.09 % (w/v) Sodium Azide
CUT&RUN Negative Control: 0.02 M Potassium Phosphate, 0.15 M NaCl, pH 7.2, 0.01 % (w/v) Sodium Azide
Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays: 20 mM Sodium Acetate pH 6.6, 20 % Ethanol -
储存液
- Sodium azide
-
注意事项
- This product contains Sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only.
-
储存条件
- 4 °C/-20 °C
-
储存方法
- Magnetic ConA Beads (Agarose) for CUT&RUN/CUT&Tag Assays ABIN6952467 must not be frozen
-
-
-
: "A distal enhancer with ETV4 binding is critical for UCP1 expression and thermogenesis in brown fat." in: Genes & development, Vol. 39, Issue 13-14, pp. 808-825, (2025) (PubMed).
-
: "A distal enhancer with ETV4 binding is critical for UCP1 expression and thermogenesis in brown fat." in: Genes & development, Vol. 39, Issue 13-14, pp. 808-825, (2025) (PubMed).
-

 (1 reference)
(1 reference) (1 validation)
(1 validation)



